GFP Bacteria
Acknowledgements
The following is based on the GFP bacteria guide from the ODIN. The tutorial is a “hello world” for bacterial engineering; it demonstrates how to culture bacteria cells, and create a human-visible change in bacteria.
A special “thank you” goes to Ash Williams for sharing his expertise, terminology, pointers, and time. He has helped me to understand more clearly what is happening in this experiment, and articulate the technical details more precisely.
Objective
The objective is to transform bacteria to express Green fluorescent protein or GFP, and fluoresce.
Procedures
The tutorial has three steps:
- Culture (12-24 hours) a colony of $\text{DH5}\alpha$ E. coli bacteria on Agar plates.
- Transform competent E. coli cells with a plasmid carrying the gene for GFP and antibiotic resistance.
- Culture (36 hours) the transformed E. coli on on LB+KAN plates, to select for the transformed cells carrying the plasmid, which are kanamycin resistant ($Kan^R$). The $Kan^R$ colonies were then screened for GFP fluorescence upon excitation with blue light.
Plasmid Visualization
The following plasmid visualization was created using SnapGene. The sequence (.fa file) is provided by the ODIN, and contains the genes for GFP and $Kan^R$.
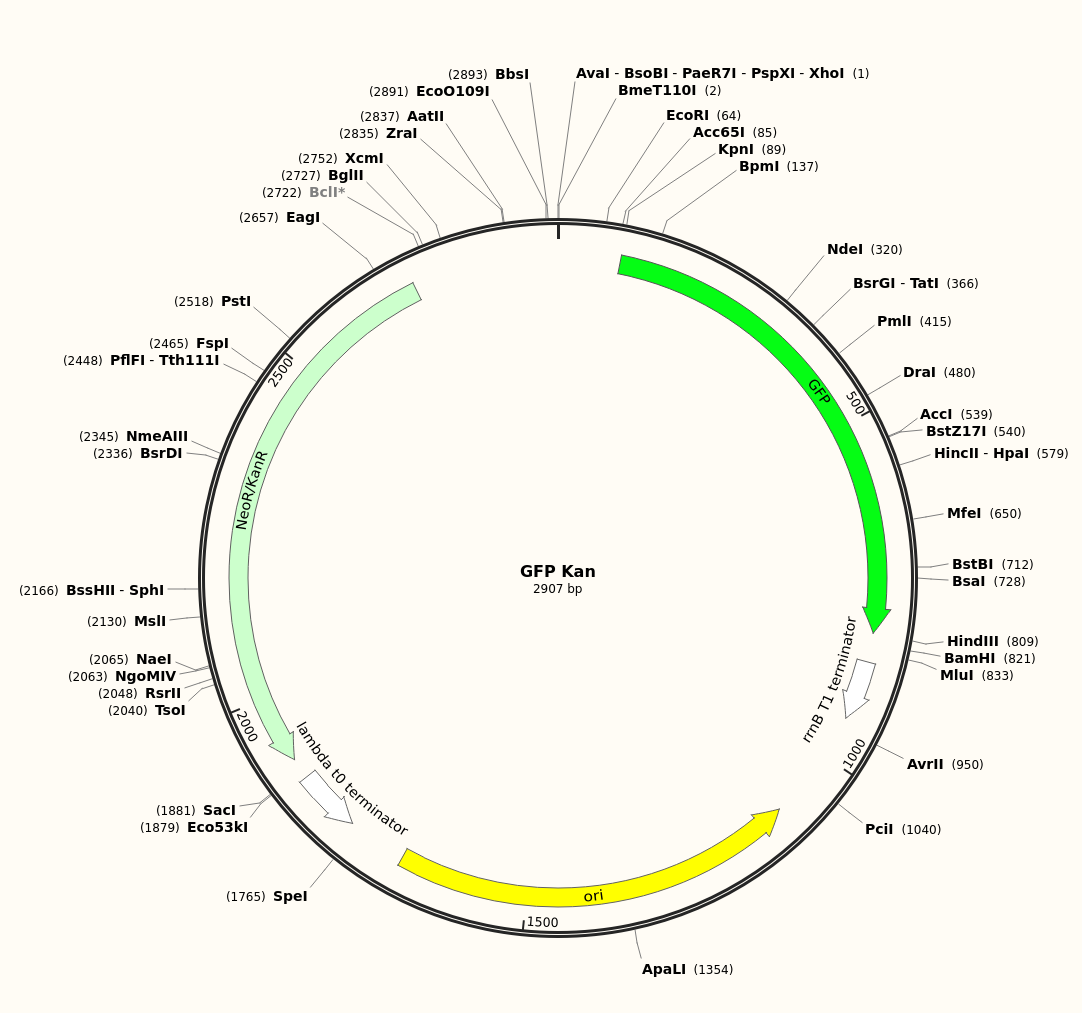
Without access to sequencing technology, the plasmid sequence above is not confirmed to be the one experimentally inserted.
Results
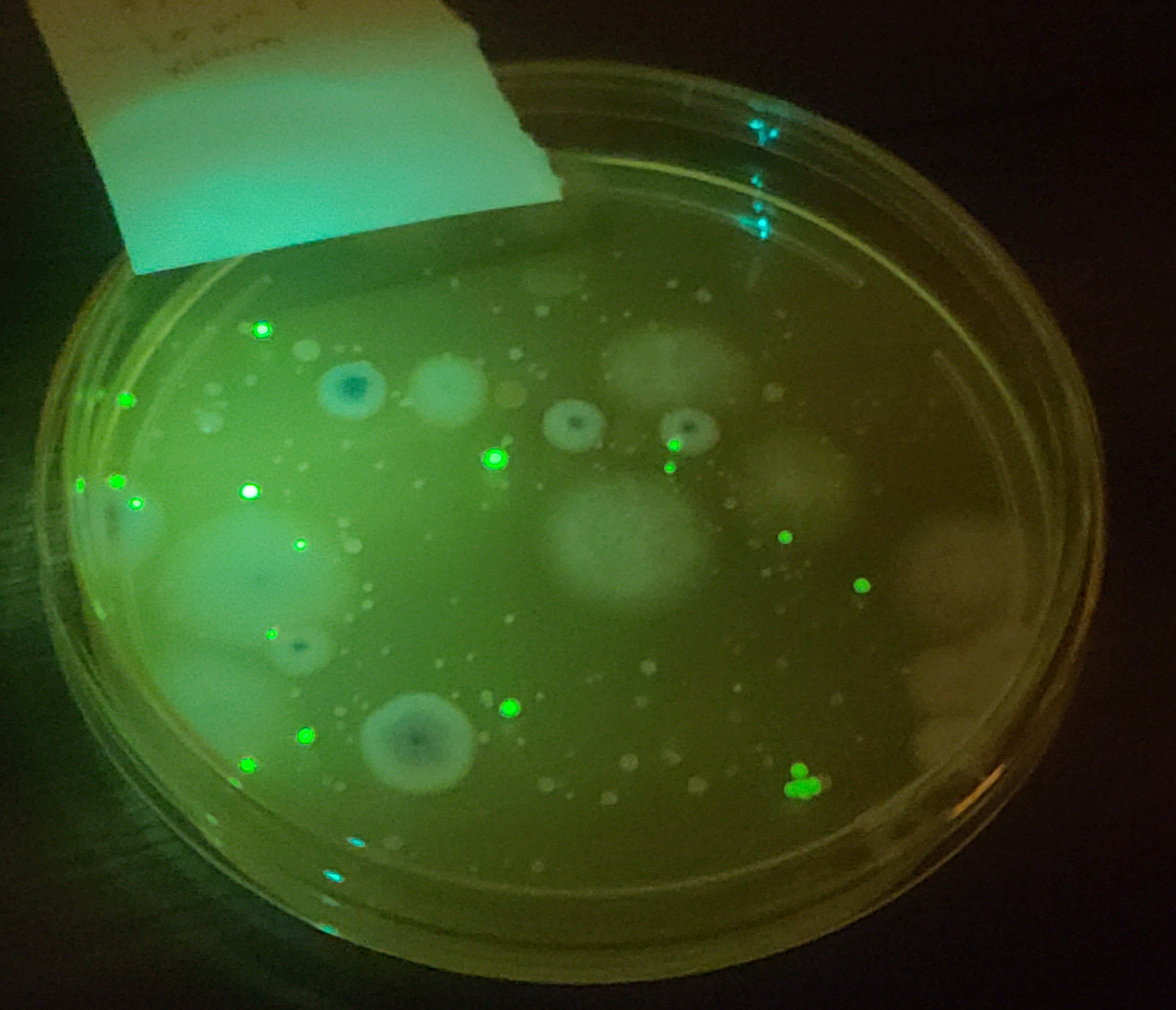
In the two images below, both full spectrum and blue light were passed through a yellow filter to produce blue light in the excitation range of GFP.
- The first plate contains colonies of transformed $Kan^R$ bacteria. The colonies are green due to fluorescence emission by excited GFP.
- The second plate applies the same imaging technique to untransformed bacteria (LB-only), which do not fluoresce.
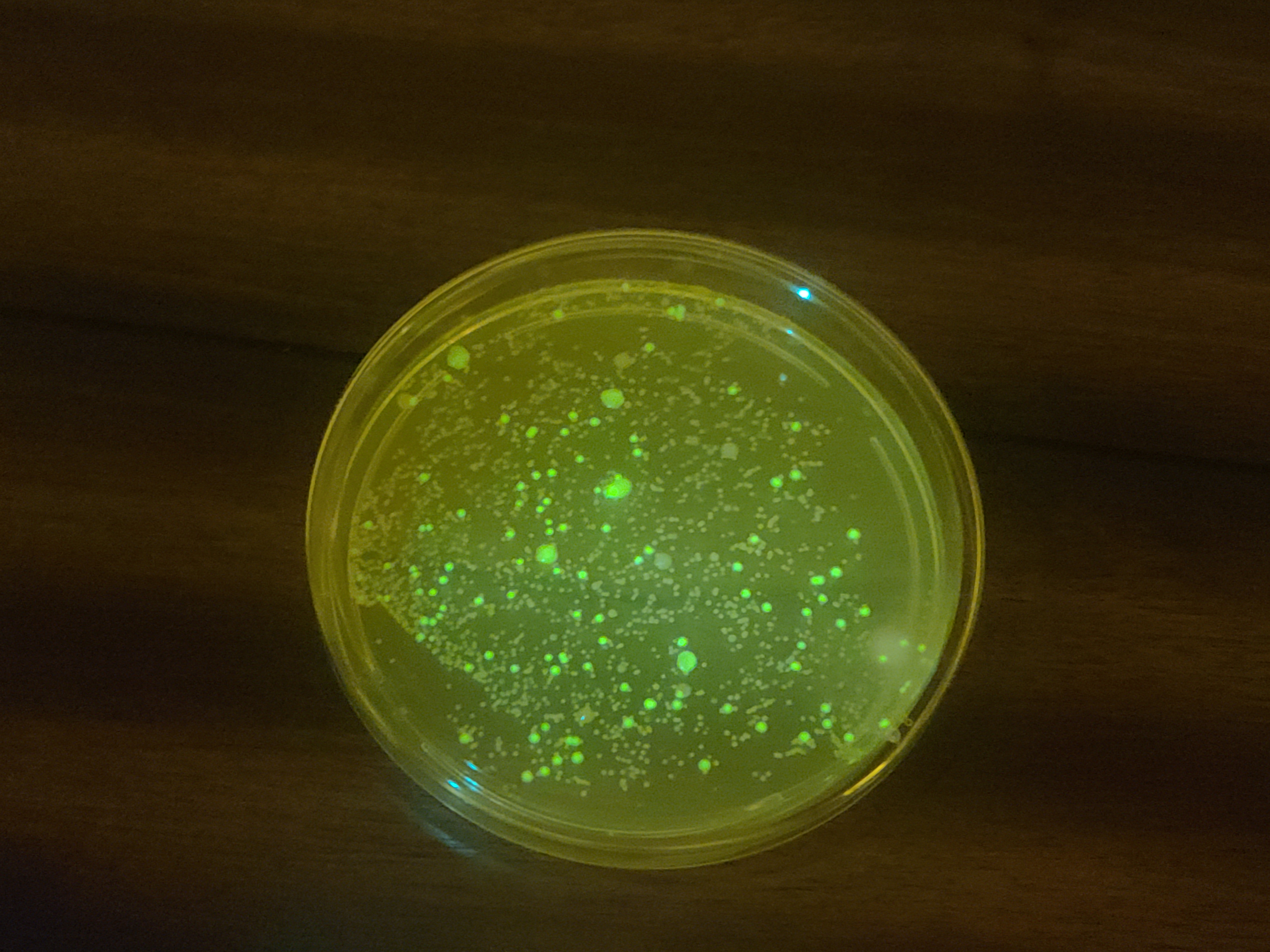 Transformed bacteria, fluorescing. Larger colonies fluoresce more, which perhaps indicates that not all GFP fluorescence is captured.
Transformed bacteria, fluorescing. Larger colonies fluoresce more, which perhaps indicates that not all GFP fluorescence is captured.
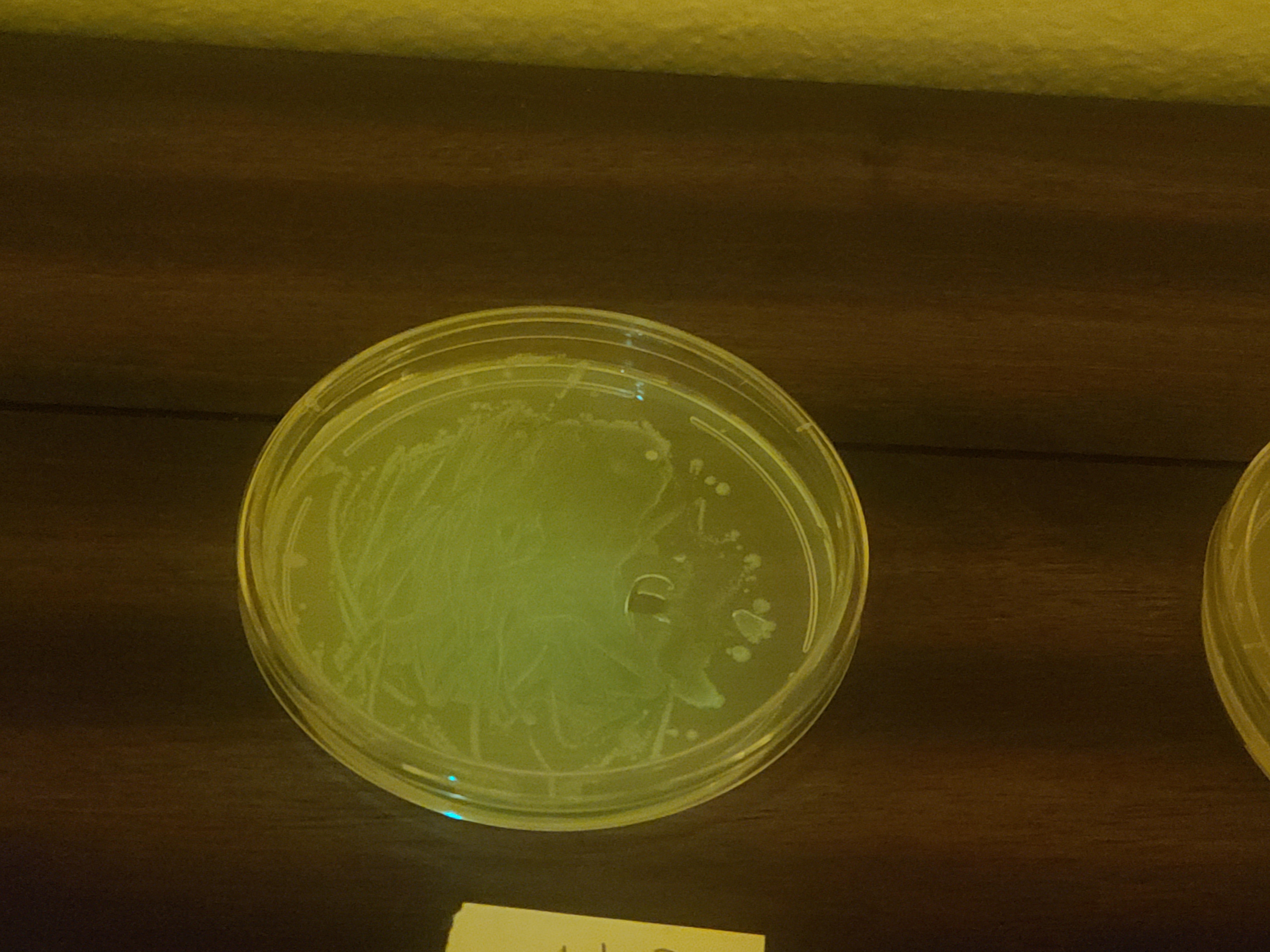 Untransformed bacteria, not fluorescing.
Untransformed bacteria, not fluorescing.
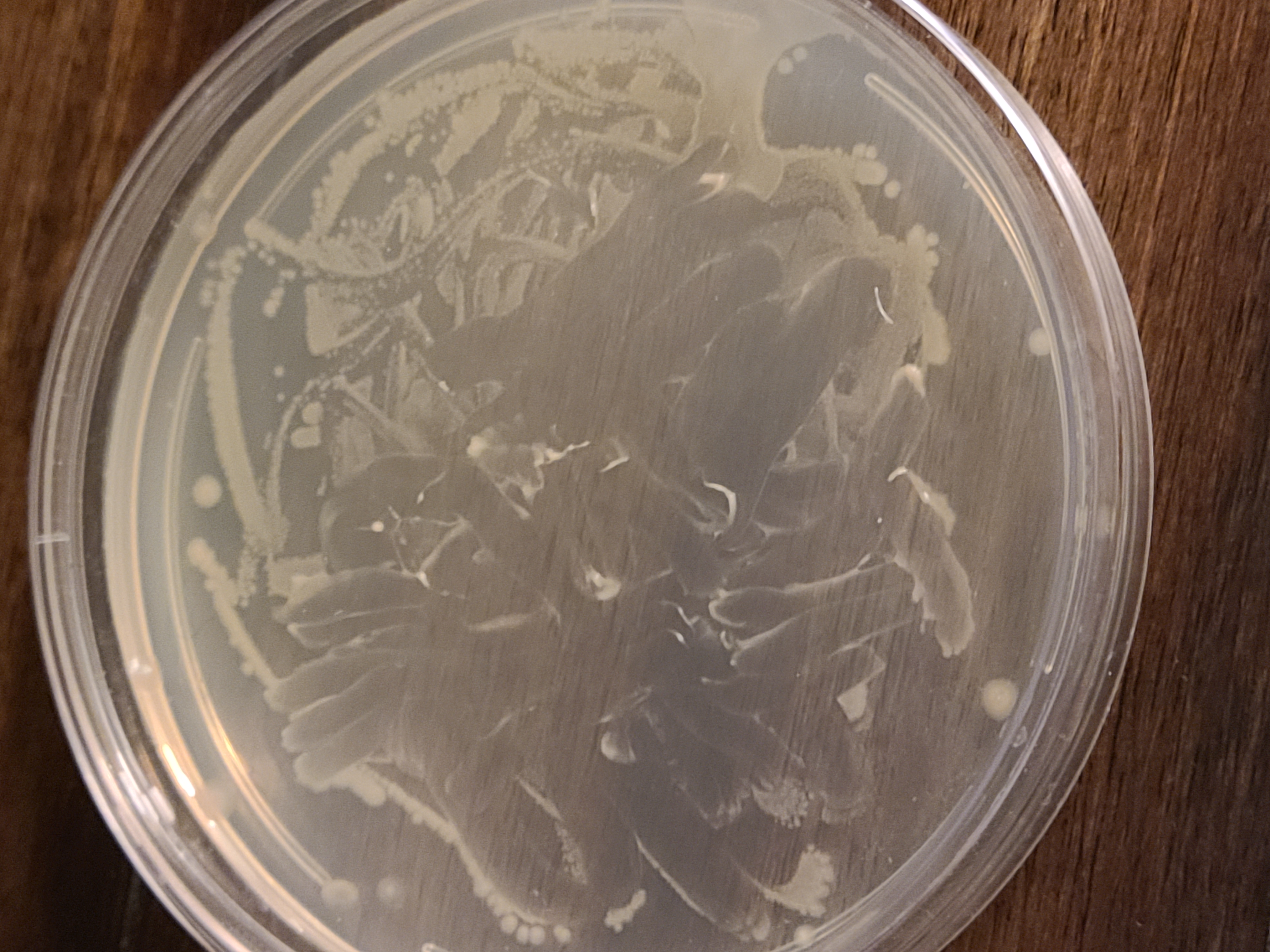
Cultures
In step (1), the initial E. coli were all similar, despite experimental variations, i.e. mixing the freeze-dried E. coli with a pipette or an inoculation loop; varying drops of the reconstituted E. coli on the plate; using inoculation loop or a pipette to add the cells to the petri dish). The cultures are depicted below.
The colonies were not necessarily isolated; some formed lawns.
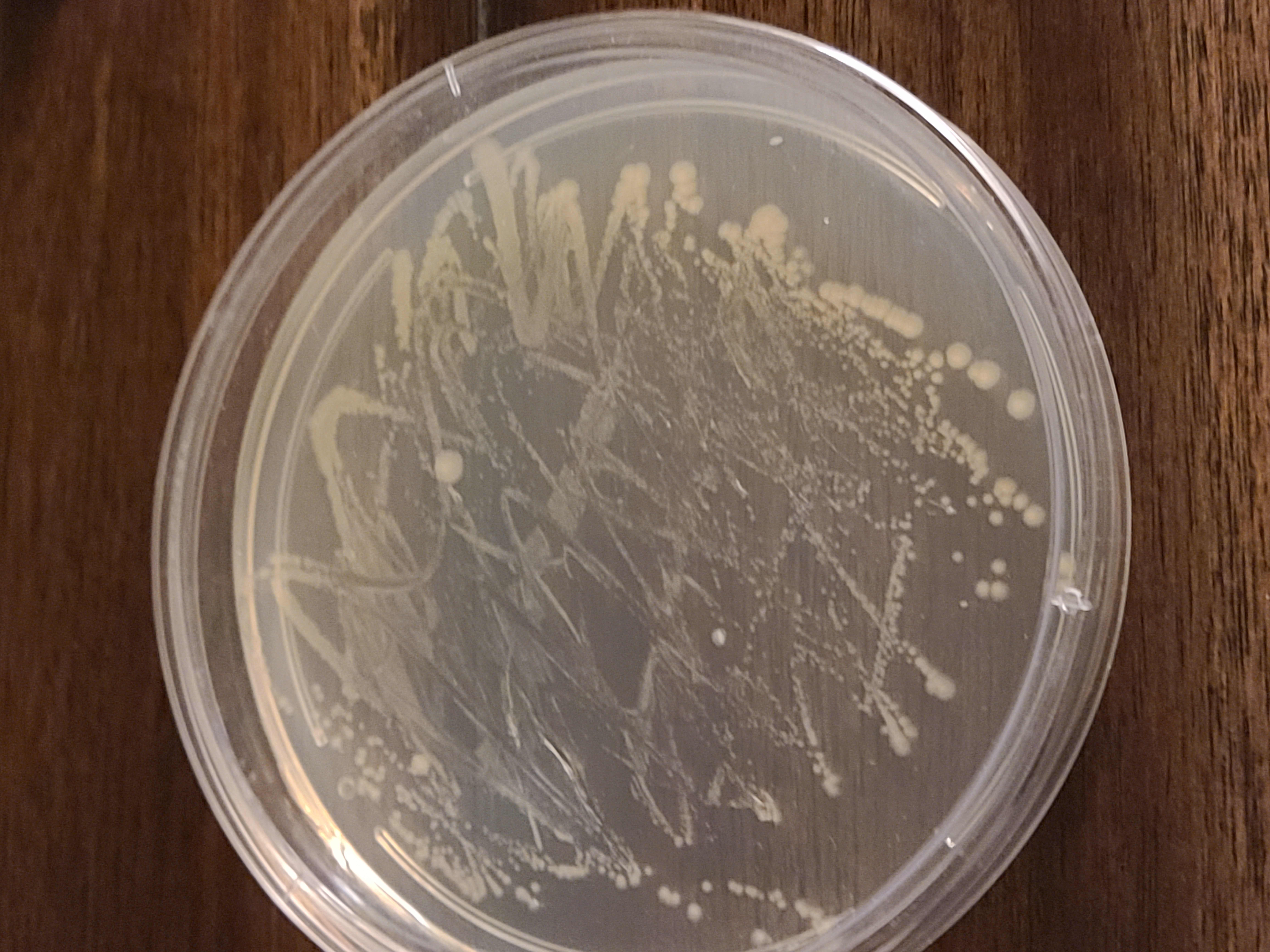
Transformations
In step (2), there were two experimental techniques to perform the transformation, varying E. coli the cell:medium density of the transformation mixture:
- “Fewer cells” variant: The small end of the inoculation loop was used to collect cells. Therefore, the transformation mixture contained less E. coli, decreasing the cell:medium ratio. A smaller ratio was thought to make the transformation more effective.
- “More cells” variant: The larger end of the inoculation loop was used to collect (a lot) of E. coli cells. Therefore, the transformation mixture contained more E. coli, thus increasing the cell:medium ratio. It was thought that a larger cell:medium ratio could make the transformation less effective. Therefore, this experimental variant relied heavily on the kanamycin in the agar to kill untransformed cells.
Cells transformed under both experimental variations were cultured.

Transformation variant 1 mixture: The transformation mixture was slighly cloudy, yet still translucent.
Transformation variant 1 produced cells screened in the Results section, which had some colonies expressing GFP.

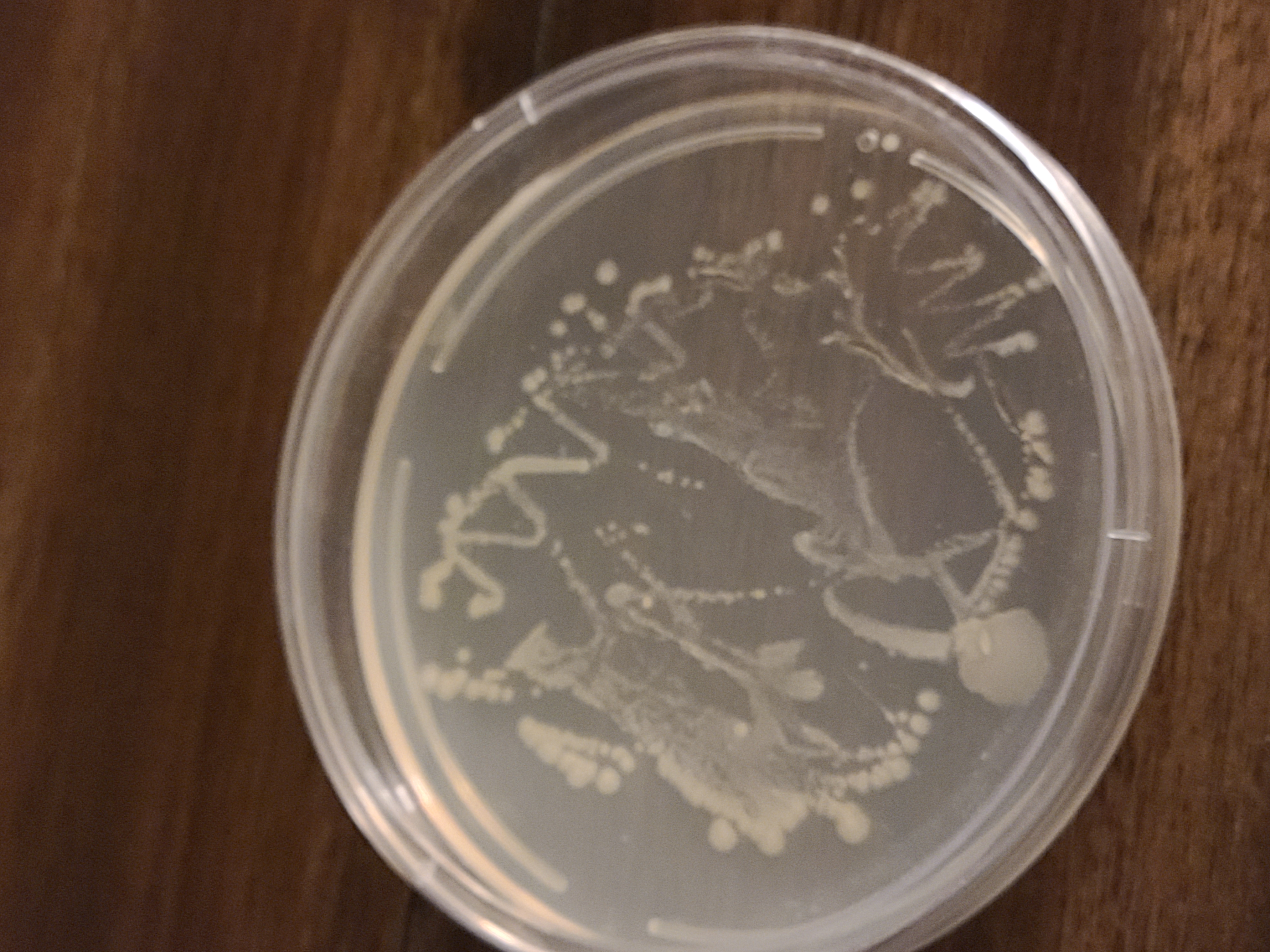
Transformation variant 2 mixture: The transformation mixture was opaque, perhaps an indication of too many E. coli cells.
Transformation variant 2 culture did produce some GFP-expressing colonies. However, the simplistic imaging technique relying on a cheap blue flashlight and a yellow, plastic screen failed to capture any fluorescence from the majority of the colonies. Either the majority of the transformations failed, or they didn’t express GFP enough for the imaging technique to capture it. This culture was also contaminated with mold, which perhaps reduced the amount that the E. coli could grow.